About
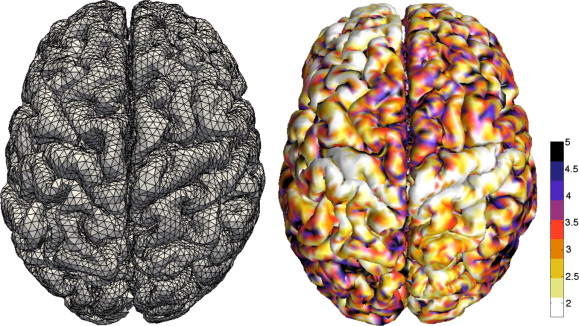
Deep learning methods have taken the MICCAI community by storm. Convolutional neural networks (CNNs) are the de-facto class of networks used in existing techniques. CNNs excel at 2D or 3D image analysis, but at the same time, there is a growing realization that not all data is organized on an ordered grid. In many applications, for example, genetics and brain imaging, data is naturally represented in a non-Euclidean space (e.g., graphs and manifolds). Moreover, the data with which the MICCAI community works contain many symmetries that can be exploited in network architectures. For this reason, geometric deep learning (GDL) has gained popularity in the medical domain for tasks including image registration, segmentation, and classification.
This tutorial seeks to draw attention to the current developments in GDL for medical image analysis. The main objective of our tutorial is to expose the vast richness of geometric structure to be found in medical image data and show how to leverage it in neural network design. We will provide a discussion on the state-of-the-art, current challenges, and opportunities. We aim to organize this tutorial as a strong complement to a workshop with the same name, that we have submitted separately. By providing this workshop in the morning, participants will get a solid foundation for the afternoon workshop.
Learning objectives
After following our tutorial, participants will be able to:
- recognize symmetries in their problems and how to exploit these.
- identify the mechanisms behind spectral and spatial graph neural networks.
- have an overview of the state-of-the-art in GDL in medical image analysis.
- identify existing challenges and opportunities of GDL when using medical data.
- critically identify the advantages of GDL and relevant components.
Program
08:00-08:05
Opening
08:05-09:00
Group convolutional networks: exploiting symmetries in medical images
Dr. Erik Bekkers, University of AmsterdamThis lecture will cover the theory behind group equivariant deep learning and show applications in medical imaging. Equivariance (if the input transforms, the output transforms in a predictable way) enables weight sharing over symmetries, enables hierarchical pattern recognition, and allows for working with geometric quantities such as (force) vectors in a principled way. I will start with an introduction to group convolutional neural networks (G-CNNs) and show that group convolutions are the most general class of equivariant linear layers. I will introduce two important directions that we are currently pursuing: (i) powerful deep learning architectures via so-called non-linear convolutions and (ii) a flexible framework to build equivariant DL architectures by parametrizing continuous convolution kernels with band-limited neural networks. Material is based on this repository.
09:00-09:55
Neural networks for learning on point clouds, graphs, and meshes
Dr. Jelmer Wolterink, University of TwenteDeep learning in medical imaging has been primarily driven by convolutional neural networks operating on 2D or 3D grids. In these, each pixel or voxel has a predefined and ordered neighborhood and consequently, convolution kernels have a fixed orientation and size. Learning on graphs requires different kinds of neural networks in which anisotropic kernels with varying numbers of neighbors can be optimized. Moreover, learning on point clouds or surface meshes – two common data representations in medical imaging – poses additional challenges. In this lecture, an overview will be given of different spectral and spatial approaches to learning on point clouds, graphs, and meshes. Applications will focus on common problems like segmentation, regression, and registration.
09:55-10:15
Coffee break
10:15-11:10
Hybrid models and hypergraph learning
Dr. Angelica Aviles-Rivero, University of CambridgeDeep supervised learning is the go-to technique for most state-of-the-art results in tasks such as classification, segmentation, and detection. However, these are heavily dependent on the availability of large and well-representative, expensive data sets. Here, we will focus on graph learning with minimal supervision. We will start by motivating graphs as a natural representation for medical data, along with the power of unlabelled data for designing robust and efficient algorithmic techniques. We will cover, from a minimal supervision perspective: i) classic models including the pros and cons of energy models and need to develop better functionals, ii) deep learning focussing on the need for higher-order relations and recent developments like hypergraphs, and iii) hybrid approaches to intertwine classic and deep learning techniques for generate robust solutions. Theory will be accompanied by real-world examples.
11:10-11:30
General Q&A and closing
Team
Dr. Jelmer Wolterink (University of Twente) works on novel deep learning methods for medical image analysis. He is the recipient of a prestigious VENI grant from the Dutch Research Council (NWO) for patient-specific aortic aneurysm modeling.
Dr. Angelica I Aviles-Rivero (University of Cambridge) centers on graph-based techniques for (bio-)medical applications, focused on novel functionals (PDEs) with carefully designed priors. Recognitions include an outstanding paper award (ICML 2020) and elected officer (SIAM SIAG/IS 2022).
Dr. Erik Bekkers (University of Amsterdam) develops deep learning methods that respect and leverage the geometric structure of (non-Euclidean) data and problems. Awards include a MICCAI Young Scientist Award (2018) and a VENI grant (NWO) for context-aware AI for medical image analysis.

Dr. Jelmer Wolterink
University of Twente
Dr. Angelica Aviles-Rivero
University of Cambridge
Dr. Erik Bekkers
University of Amsterdam